Table of Contents
A description on how secondary structures are decomposed into individual loops to eventually evaluate their stability in terms of free energy.
Secondary Structure Loop Decomposition
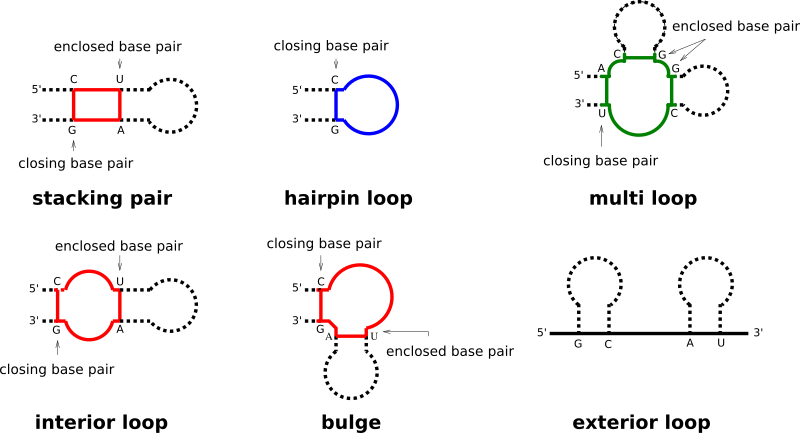
Each base pair in a secondary structure closes a loop, thereby directly enclosing unpaired nucleotides, and/or further base pairs. Our implementation distinguishes four basic types of loops:
- hairpin loops
- interior loops
- multibranch loops
- exterior loop
While the exterior loop is a special case without a closing pair, the other loops are determined by the number of base pairs involved in the loop formation, i.e. hairpin loops are 1-loops, since only a single base pair delimits the loop. interior loops are 2-loops due to their enclosing, and enclosed base pair. All loops where more than two base pairs are involved, are termed multibranch loops.
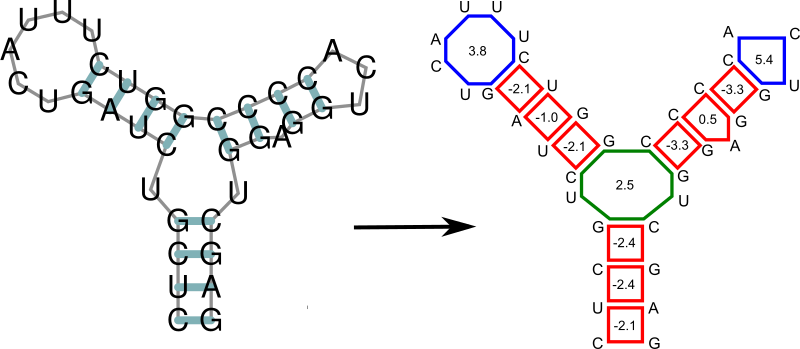
Any secondary structure can be decomposed into its loops. Each of the loops then can be scored in terms of free energy, and the free energy of an entire secondary structure is simply the sum of free energies of its loops.
Free Energy Evaluation API
While we implement some functions that decompose a secondary structure into its individual loops, the majority of methods provided in are dedicated to free energy evaluation. The corresponding modules are:
Free Energy Parameters
For secondary structure free energy evaluation we usually utilize the set of Nearest Neighbor Parameters also used in other software, such as UNAfold and RNAstructure. While the RNAlib already contains a compiled-in set of the latest Turner 2004 Free Energy Parameters, we defined a file format that allows to change these parameters at runtime. The ViennaRNA Package already comes with a set of parameter files containing
- Turner 1999 RNA parameters
- Mathews 1999 DNA parameters
- Andronescu 2007 RNA parameters
- Mathews 2004 DNA parameters
Free Energy Parameters Modification API
Fine-tuning of the Energy Evaluation Model
Go to the next Chapter "Secondary Structure Folding Grammar"
