Utilities
We also ship a number of small utilities, many of them to manipulate the PostScript files produced by the structure prediction programs RNAfold and RNAalifold.
Most of the Perl 5 utilities contain embedded pod documentation. Type e.g.
perldoc relplot.pl
for detailed instructions.
Available Tools
Tool Name |
Description |
|---|---|
ct2db |
Produce dot bracket notation of an RNA secondary structure
given as mfold
.ct file |
b2mt.pl |
Produce a mountain representation of a secondary structure
from it’s dot-bracket notation, as produced by RNAfold.
Output consists of simple
x y data suitable as input to aplotting program. The mountain representation is a xy plot with
sequence position on the x-axis and the number of base pairs
enclosing that position on the y-axis.
Example: RNAfold < my.seq | b2mt.pl | xmgrace -pipe
|
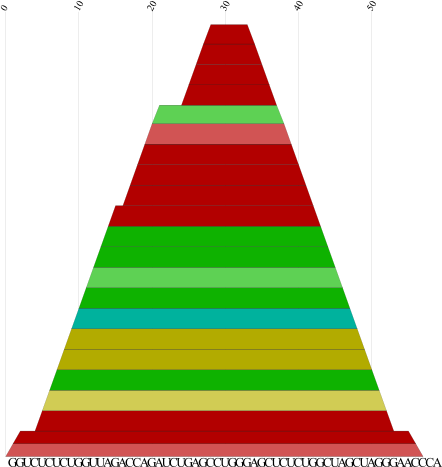
cmount.pl |
Produce a PostScript mountain plot from a color dot plot as
created by RNAalifold -p or
alidot -p. Each base pairis represented by a trapez whose color encodes the number of
consistent and compensatory mutations supporting that pair:
Red marks pairs with no sequence variation; ochre, green, turquoise,
blue, and violet mark pairs with 2,3,4,5,6 different types of pairs,
respectively.
Example: cmount.pl alidot.ps > cmount.ps

|
coloraln.pl |
Reads a sequence alignment in
CLUSTAL format and a consensussecondary structure (which it extracts from a secondary structure
plot as produced by RNAalifold), and produces a postscript figure
of the alignment annotated using the consensus structure, coloring
base pair using the same color scheme as
cmount.pl, RNAalifoldand
alidot.Example: coloraln.pl -s alirna.ps file.aln > coloraln.ps

|
colorrna.pl |
Reads a consensus secondary structure plot and a color dot plot
as produced by RNAalifold -p, and writes a new secondary
structure plot in which base pairs a colored using the color
information from the dot plot.
Example: colorrna.pl alirna.ps alidot.ps > colorRNA.ps
|
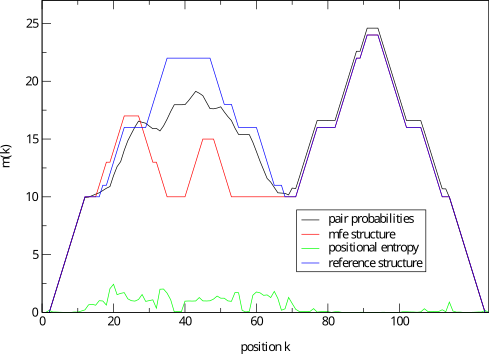
mountain.pl |
Similar to
b2mt.pl, but produces a mountain plot from thepair probabilities contained in a PostScript dot plot. It write
3 sets of x y data, suitable as input for a plot program. The
first two sets containing the mountain representation from pair
probabilities and MFE structure, the third set is the
“positional entropy” a measure of structural well-definedness.
Example: mountain.pl dot.ps | xmgrace -pipe
|
refold.pl |
Refold using consensus structure as constraint |
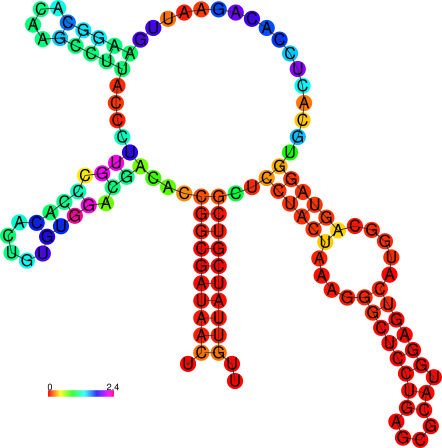
relplot.pl |
Reads a postscript secondary structure plot and a dot plot
containing pair probabilities as produced by RNAfold -p,
and produces a new structure plot, color annotated with reliability
information in the form of either pair probabilities or positional
entropy (default).
Example: relplot.pl foo_ss.ps foo_dp.ps > foo_rss.ps
|
rotate_ss.pl |
Reads a postscript secondary structure plot as produced by
RNAfold and produces a new rotated and/or mirrored
structure plot.
Example: rotate_ss.pl -a 30 -m foo_ss.ps > foo_new_ss.ps
|
switch.pl |
Design sequences that can adopt two different structure, i.e.
design RNA switches. The program will sample the set of sequences
compatible with two input structures in order to find sequences with
desired thermodynamic and kinetic properties. In particular it is
possible to specify two different temperatures such that structure
1 is favored at T1 and structure 2 at T2 to design temperature
sensitive switches (RNA thermometers). The desired height of the
energy barrier separating the two structures, thus determining
the refolding time between meta-stable states.
|
RNAdesign.pl |
Flexible design of multi-stable RNA molecules. An initially random
sequence is iteratively mutated and evaluated according to an
objective function (see Option:
--optfun). Whenever a betterscoring sequence has been found, the mutation is accepted, the
algorithm terminates once a local minimum is found. This script
makes heavy use of the
RNA::Design sub-package that comeswith the ViennaRNA Package
Perl 5 interface. |