bamPEFragmentSize¶
This tool calculates the fragment sizes for read pairs given a BAM file from paired-end sequencing.Several regions are sampled depending on the size of the genome and number of processors to estimate thesummary statistics on the fragment lengths. Properly paired reads are preferred for computation, i.e., it will only use discordant pairs if no concordant alignments overlap with a given region. The default setting simply prints the summary statistics to the screen.
usage: bamPEFragmentSize [-h] [--bamfiles bam files [bam files ...]] [--histogram FILE]
[--plotFileFormat FILETYPE] [--numberOfProcessors INT]
[--samplesLabel SAMPLESLABEL [SAMPLESLABEL ...]] [--plotTitle PLOTTITLE]
[--maxFragmentLength MAXFRAGMENTLENGTH] [--logScale] [--binSize INT]
[--distanceBetweenBins INT] [--blackListFileName BED file] [--table FILE]
[--outRawFragmentLengths FILE] [--verbose] [--version]
Named Arguments¶
- --bamfiles, -b
List of BAM files to process
- --histogram, -hist, -o
Save a .png file with a histogram of the fragment length distribution.
- --plotFileFormat
Possible choices: png, pdf, svg, eps, plotly
Image format type. If given, this option overrides the image format based on the plotFile ending. The available options are: png, eps, pdf, svg and plotly.
- --numberOfProcessors, -p
Number of processors to use. The default is to use 1. (Default: 1)
- --samplesLabel
Labels for the samples plotted. The default is to use the file name of the sample. The sample labels should be separated by spaces and quoted if a label itselfcontains a space E.g. –samplesLabel label-1 “label 2”
- --plotTitle, -T
Title of the plot, to be printed on top of the generated image. Leave blank for no title. (Default: )
- --maxFragmentLength
The maximum fragment length in the histogram. A value of 0 (the default) indicates to use twice the mean fragment length. (Default: 0)
- --logScale
Plot on the log scale
- --binSize, -bs
Length in bases of the window used to sample the genome. (Default: 1000)
- --distanceBetweenBins, -n
To reduce the computation time, not every possible genomic bin is sampled. This option allows you to set the distance between bins actually sampled from. Larger numbers are sufficient for high coverage samples, while smaller values are useful for lower coverage samples. Note that if you specify a value that results in too few (<1000) reads sampled, the value will be decreased. (Default: 1000000)
- --blackListFileName, -bl
A BED file containing regions that should be excluded from all analyses. Currently this works by rejecting genomic chunks that happen to overlap an entry. Consequently, for BAM files, if a read partially overlaps a blacklisted region or a fragment spans over it, then the read/fragment might still be considered.
- --table
In addition to printing read and fragment length metrics to the screen, write them to the given file in tabular format.
- --outRawFragmentLengths
Save the fragment (or read if the input is single-end) length and their associated number of occurrences to a tab-separated file. Columns are length, number of occurrences, and the sample label.
- --verbose
Set if processing data messages are wanted.
- --version
show program’s version number and exit
Example usage¶
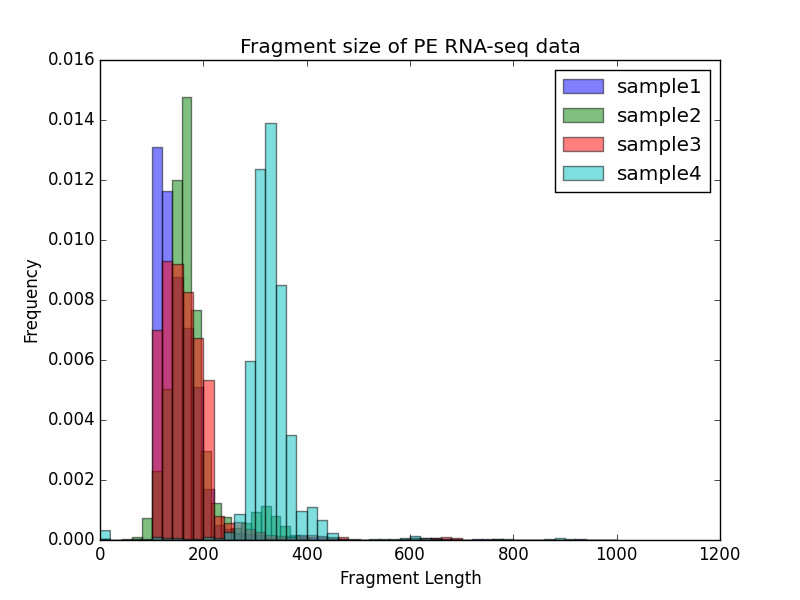
$ deepTools2.0/bin/bamPEFragmentSize \
-hist fragmentSize.png \
-T "Fragment size of PE RNA-seq data" \
--maxFragmentLength 1000 \
-b testFiles/RNAseq_sample1.bam testFiles/RNAseq_sample2.bam \
testFiles/RNAseq_sample3.bam testFiles/RNAseq_sample4.bam \
-samplesLabel sample1 sample2 sample3 sample4
## Output
BAM file : testFiles/RNAseq_sample1.bam
Sample size: 10815
Fragment lengths:
Min.: 0.0
1st Qu.: 311.0
Mean: 8960.68987517
Median: 331.0
3rd Qu.: 362.0
Max.: 53574842.0
Std: 572421.46625
Read lengths:
Min.: 20.0
1st Qu.: 101.0
Mean: 99.1621821544
Median: 101.0
3rd Qu.: 101.0
Max.: 101.0
Std: 9.16567362755
BAM file : testFiles/RNAseq_sample2.bam
Sample size: 6771
Fragment lengths:
Min.: 43.0
1st Qu.: 148.0
Mean: 176.465071629
Median: 164.0
3rd Qu.: 185.0
Max.: 500.0
Std: 53.733877263
......(output truncated)
If the --table option is specified, the summary statistics are additionally printed in a tabular format:
Frag. Len. Min. Frag. Len. 1st. Qu. Frag. Len. Mean Frag. Len. Median Frag. Len. 3rd Qu. Frag. Len. Max Frag. Len. Std. Read Len. Min. Read Len. 1st. Qu. Read Len. Mean Read Len. Median Read Len. 3rd Qu. Read Len. Max Read Len. Std.
bowtie2 test1.bam 241.0 241.5 244.666666667 242.0 246.5 251.0 4.49691252108 251.0 251.0 251.0 251.0 251.0 251.0 0.0
If the --outRawFragmentLengths option is provided, another history item will be produced, containing the raw data underlying the histogram. It has the following format:
#bamPEFragmentSize
Size Occurrences Sample
241 1 bowtie2 test1.bam
242 1 bowtie2 test1.bam
251 1 bowtie2 test1.bam
The “Size” is the fragment (or read, for single-end datasets) size and “Occurrences” are the number of times reads/fragments with that length were observed. For easing downstream processing, the sample name is a lso included on each row.