Grantham (1974) Amino-Acid chemical distance. More...
#include <Bpp/Seq/AlphabetIndex/GranthamAAChemicalDistance.h>
 Inheritance diagram for bpp::GranthamAAChemicalDistance:
Inheritance diagram for bpp::GranthamAAChemicalDistance: Collaboration diagram for bpp::GranthamAAChemicalDistance:
Collaboration diagram for bpp::GranthamAAChemicalDistance:Public Member Functions | |
| GranthamAAChemicalDistance () | |
| GranthamAAChemicalDistance (const GranthamAAChemicalDistance &gd) | |
| GranthamAAChemicalDistance & | operator= (const GranthamAAChemicalDistance &gd) |
| virtual | ~GranthamAAChemicalDistance () |
| void | setSymmetric (bool yn) |
| bool | isSymmetric () const |
| void | setPC1Sign (bool yn) |
| The sign of the distance is computed using the coordinate on the first axis of a principal component analysis with the 3 elementary properties (Volume, Polarity, Composition). Otherwise, use the default arbitrary sign. Using this option will lead isSymmetric to return false. More... | |
Methods from the AlphabetIndex2 interface. | |
| double | getIndex (int state1, int state2) const throw (BadIntException) |
| Get the index associated to a pair of states. More... | |
| double | getIndex (const std::string &state1, const std::string &state2) const throw (BadCharException) |
| Get the index associated to a pair of states. More... | |
| const Alphabet * | getAlphabet () const |
| Get the alphabet associated to this index. More... | |
| GranthamAAChemicalDistance * | clone () const |
| Matrix< double > * | getIndexMatrix () const |
Static Public Attributes | |
| static short int | SIGN_ARBITRARY = 1 |
| static short int | SIGN_PC1 = 2 |
| static short int | SIGN_NONE = 0 |
Private Attributes | |
| LinearMatrix< double > | distanceMatrix_ |
| LinearMatrix< double > | signMatrix_ |
| const ProteicAlphabet * | alpha_ |
| short int | sign_ |
Detailed Description
Grantham (1974) Amino-Acid chemical distance.
Two kinds of matrix can be built:
- a symmetric one, with
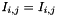 ,
, - or a non-symmetric one, with
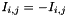 . In the second case, which one of the two entries between
. In the second case, which one of the two entries between 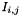 and
and 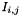 is positive is arbitrary by default. It is also possible to use the coordinate on a principal component analysis between the elementary propoerties of the distance instead (setPC1Sign(true)). The following R code was use in order to get those signs: library(seqinr)data(aaindex)data<-data.frame(composition=aaindex[["GRAR740101"]]$I,polarity=aaindex[["GRAR740102"]]$I,volume=aaindex[["GRAR740103"]]$I)library(ade4)pca<-dudi.pca(data)plot(pca$li[, 1:2], type="n")text(pca$li[, 1:2], rownames(data))s.corcircle(pca$co)layout(matrix(1:3,nrow=1))a1<-pca$li[,1]; names(a1)<-rownames(data); dotchart(sort(a1))a2<-pca$li[,2]; names(a2)<-rownames(data); dotchart(sort(a2))a3<-pca$li[,3]; names(a3)<-rownames(data); dotchart(sort(a3))x<-pca$li[,1] #Contains the coordinates on the first axis.m<-matrix(nrow=20, ncol=20)for(i in 1:length(x))for(j in 1:length(x))m[i,j]<-sign(x[j] - x[i])
is positive is arbitrary by default. It is also possible to use the coordinate on a principal component analysis between the elementary propoerties of the distance instead (setPC1Sign(true)). The following R code was use in order to get those signs: library(seqinr)data(aaindex)data<-data.frame(composition=aaindex[["GRAR740101"]]$I,polarity=aaindex[["GRAR740102"]]$I,volume=aaindex[["GRAR740103"]]$I)library(ade4)pca<-dudi.pca(data)plot(pca$li[, 1:2], type="n")text(pca$li[, 1:2], rownames(data))s.corcircle(pca$co)layout(matrix(1:3,nrow=1))a1<-pca$li[,1]; names(a1)<-rownames(data); dotchart(sort(a1))a2<-pca$li[,2]; names(a2)<-rownames(data); dotchart(sort(a2))a3<-pca$li[,3]; names(a3)<-rownames(data); dotchart(sort(a3))x<-pca$li[,1] #Contains the coordinates on the first axis.m<-matrix(nrow=20, ncol=20)for(i in 1:length(x))for(j in 1:length(x))m[i,j]<-sign(x[j] - x[i])
Reference: Grantham, R. Amino acid difference formula to help explain protein evolution Science 185, 862-864 (1974)
Data from AAIndex2 database, Accession Number GRAR740104.
Definition at line 96 of file GranthamAAChemicalDistance.h.
Constructor & Destructor Documentation
◆ GranthamAAChemicalDistance() [1/2]
| GranthamAAChemicalDistance::GranthamAAChemicalDistance | ( | ) |
Definition at line 53 of file GranthamAAChemicalDistance.cpp.
Referenced by clone().
◆ GranthamAAChemicalDistance() [2/2]
|
inline |
Definition at line 108 of file GranthamAAChemicalDistance.h.
◆ ~GranthamAAChemicalDistance()
|
virtual |
Definition at line 62 of file GranthamAAChemicalDistance.cpp.
Member Function Documentation
◆ clone()
|
inlinevirtual |
Implements bpp::AlphabetIndex2.
Definition at line 135 of file GranthamAAChemicalDistance.h.
References GranthamAAChemicalDistance().
◆ getAlphabet()
|
inlinevirtual |
Get the alphabet associated to this index.
- Returns
- Alphabet The alphabet associated to this index.
Implements bpp::AlphabetIndex2.
Definition at line 134 of file GranthamAAChemicalDistance.h.
References alpha_.
◆ getIndex() [1/2]
|
virtual | ||||||||||||||||||||
Get the index associated to a pair of states.
- Parameters
-
state1 First state to consider, as a int value. state2 Second state to consider, as a int value.
- Returns
- The index associated to the pair of states.
Implements bpp::AlphabetIndex2.
Definition at line 64 of file GranthamAAChemicalDistance.cpp.
◆ getIndex() [2/2]
|
virtual | ||||||||||||||||||||
Get the index associated to a pair of states.
- Parameters
-
state1 First state to consider, as a string value. state2 Second state to consider, as a string value.
- Returns
- The index associated to the pair of states.
Implements bpp::AlphabetIndex2.
Definition at line 77 of file GranthamAAChemicalDistance.cpp.
◆ getIndexMatrix()
|
virtual |
- Returns
- A matrix object with all indices.
Implements bpp::AlphabetIndex2.
Definition at line 83 of file GranthamAAChemicalDistance.cpp.
References distanceMatrix_, sign_, SIGN_NONE, SIGN_PC1, and signMatrix_.
◆ isSymmetric()
|
inlinevirtual |
- Returns
- True if the index is symatric (that is, index(i,j) == index(j, i)).
Implements bpp::AlphabetIndex2.
Definition at line 141 of file GranthamAAChemicalDistance.h.
◆ operator=()
|
inline |
Definition at line 115 of file GranthamAAChemicalDistance.h.
References alpha_, distanceMatrix_, sign_, and signMatrix_.
◆ setPC1Sign()
|
inline |
The sign of the distance is computed using the coordinate on the first axis of a principal component analysis with the 3 elementary properties (Volume, Polarity, Composition). Otherwise, use the default arbitrary sign. Using this option will lead isSymmetric to return false.
- Parameters
-
yn Tell is the PC1-based sign should be used instead of the arbitrary one.
Definition at line 149 of file GranthamAAChemicalDistance.h.
References sign_, SIGN_ARBITRARY, and SIGN_PC1.
Referenced by bpp::BppOAlphabetIndex2Format::read().
◆ setSymmetric()
|
inline |
Definition at line 140 of file GranthamAAChemicalDistance.h.
References sign_, SIGN_ARBITRARY, and SIGN_NONE.
Referenced by bpp::BppOAlphabetIndex2Format::read().
Member Data Documentation
◆ alpha_
|
private |
Definition at line 102 of file GranthamAAChemicalDistance.h.
Referenced by getAlphabet(), and operator=().
◆ distanceMatrix_
|
private |
Definition at line 100 of file GranthamAAChemicalDistance.h.
Referenced by getIndexMatrix(), and operator=().
◆ sign_
|
private |
Definition at line 103 of file GranthamAAChemicalDistance.h.
Referenced by getIndexMatrix(), isSymmetric(), operator=(), setPC1Sign(), and setSymmetric().
◆ SIGN_ARBITRARY
|
static |
Definition at line 151 of file GranthamAAChemicalDistance.h.
Referenced by setPC1Sign(), and setSymmetric().
◆ SIGN_NONE
|
static |
Definition at line 153 of file GranthamAAChemicalDistance.h.
Referenced by getIndexMatrix(), isSymmetric(), and setSymmetric().
◆ SIGN_PC1
|
static |
Definition at line 152 of file GranthamAAChemicalDistance.h.
Referenced by getIndexMatrix(), and setPC1Sign().
◆ signMatrix_
|
private |
Definition at line 101 of file GranthamAAChemicalDistance.h.
Referenced by getIndexMatrix(), and operator=().
The documentation for this class was generated from the following files:
- Bpp/Seq/AlphabetIndex/GranthamAAChemicalDistance.h
- Bpp/Seq/AlphabetIndex/GranthamAAChemicalDistance.cpp