The Goldman and Yang (1994) substitution model for codons. More...
#include <Bpp/Phyl/Model/Codon/GY94.h>
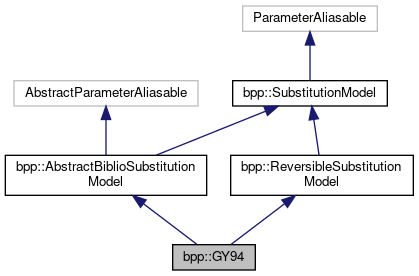
 Inheritance diagram for bpp::GY94:
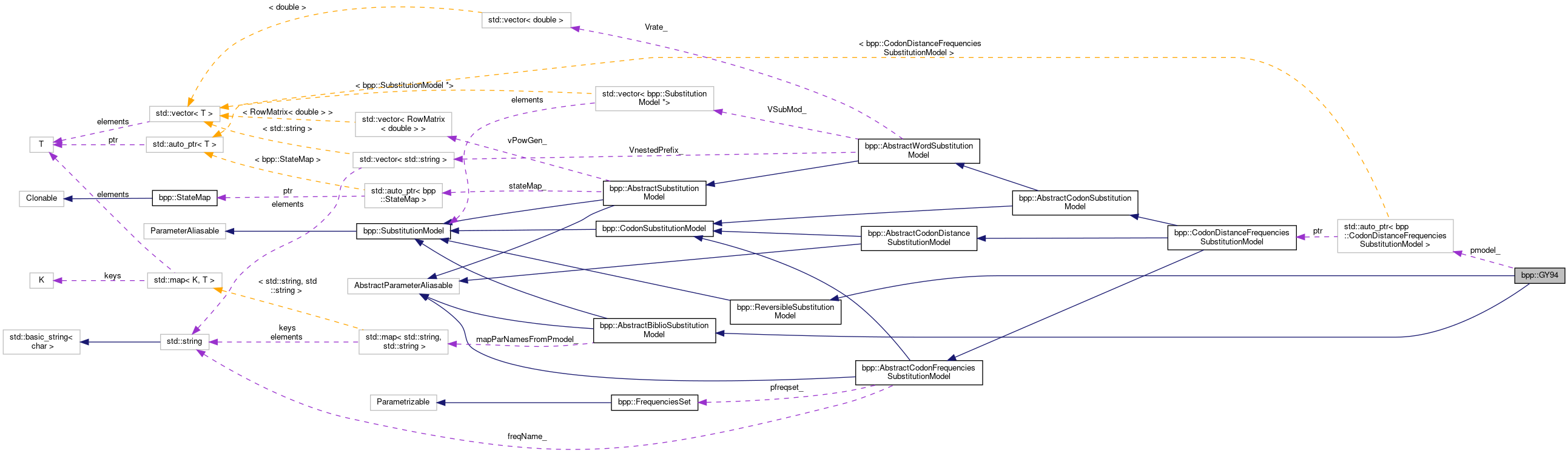
Inheritance diagram for bpp::GY94: Collaboration diagram for bpp::GY94:
Collaboration diagram for bpp::GY94:Public Member Functions | |
| GY94 (const GeneticCode *gc, FrequenciesSet *codonFreqs) | |
| ~GY94 () | |
| GY94 (const GY94 &gy94) | |
| GY94 & | operator= (const GY94 &gy94) |
| GY94 * | clone () const |
| std::string | getName () const |
| Get the name of the model. More... | |
| const SubstitutionModel & | getModel () const |
| const std::vector< int > & | getAlphabetStates () const |
| const StateMap & | getStateMap () const |
| int | getAlphabetStateAsInt (size_t i) const |
| std::string | getAlphabetStateAsChar (size_t i) const |
| std::vector< size_t > | getModelStates (int code) const |
| Get the state in the model corresponding to a particular state in the alphabet. More... | |
| std::vector< size_t > | getModelStates (const std::string &code) const |
| Get the state in the model corresponding to a particular state in the alphabet. More... | |
| virtual double | freq (size_t i) const |
| virtual double | Qij (size_t i, size_t j) const |
| virtual double | Pij_t (size_t i, size_t j, double t) const |
| virtual double | dPij_dt (size_t i, size_t j, double t) const |
| virtual double | d2Pij_dt2 (size_t i, size_t j, double t) const |
| virtual const Vdouble & | getFrequencies () const |
| const Matrix< double > & | getGenerator () const |
| const Matrix< double > & | getExchangeabilityMatrix () const |
| double | Sij (size_t i, size_t j) const |
| const Matrix< double > & | getPij_t (double t) const |
| const Matrix< double > & | getdPij_dt (double t) const |
| const Matrix< double > & | getd2Pij_dt2 (double t) const |
| void | enableEigenDecomposition (bool yn) |
| Set if eigenValues and Vectors must be computed. More... | |
| bool | enableEigenDecomposition () |
| Tell if eigenValues and Vectors must be computed. More... | |
| bool | isDiagonalizable () const |
| bool | isNonSingular () const |
| const Vdouble & | getEigenValues () const |
| const Vdouble & | getIEigenValues () const |
| const Matrix< double > & | getRowLeftEigenVectors () const |
| const Matrix< double > & | getColumnRightEigenVectors () const |
| double | getRate () const |
| Get the rate. More... | |
| void | setRate (double rate) |
| Set the rate of the model (must be positive). More... | |
| void | addRateParameter () |
| void | setFreqFromData (const SequenceContainer &data, double pseudoCount=0) |
| Set equilibrium frequencies equal to the frequencies estimated from the data. More... | |
| void | setFreq (std::map< int, double > &frequ) |
| Set equilibrium frequencies. More... | |
| const Alphabet * | getAlphabet () const |
| size_t | getNumberOfStates () const |
| Get the number of states. More... | |
| double | getInitValue (size_t i, int state) const throw (BadIntException) |
| const FrequenciesSet * | getFrequenciesSet () const |
| If the model owns a FrequenciesSet, returns a pointer to it, otherwise return 0. More... | |
| virtual void | fireParameterChanged (const ParameterList ¶meters) |
| Tells the model that a parameter value has changed. More... | |
| double | getScale () const |
| Get the scalar product of diagonal elements of the generator and the frequencies vector. If the generator is normalized, then scale=1. Otherwise each element must be multiplied by 1/scale. More... | |
| void | setScale (double scale) |
| Multiplies the current generator by the given scale. More... | |
Protected Member Functions | |
| virtual void | updateMatrices () |
Protected Attributes | |
| std::map< std::string, std::string > | mapParNamesFromPmodel_ |
| Tools to make the link between the Parameters of the object and those of pmixmodel_. More... | |
| ParameterList | lParPmodel_ |
Private Member Functions | |
| SubstitutionModel & | getModel () |
Private Attributes | |
| GranthamAAChemicalDistance | gacd_ |
| std::auto_ptr< CodonDistanceFrequenciesSubstitutionModel > | pmodel_ |
Detailed Description
The Goldman and Yang (1994) substitution model for codons.
This model has one rate of transitions and one rate of transversion. It also allows distinct equilibrium frequencies between codons. A multiplicative factor accounts for the selective restraints at the amino acid level. This factor applies on the distance 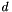 between amino acids given by Grantham (1974).
between amino acids given by Grantham (1974).
For codons 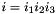 and
and 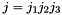 , the generator term
, the generator term 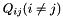 is:
is:
0 if 2 or 3 of the pair 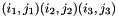 are different.
are different.
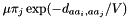 if exactly 1 of the pairs
if exactly 1 of the pairs 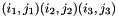 is different, and that difference is a transversion.
is different, and that difference is a transversion.
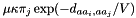 if exactly 1 of the pairs
if exactly 1 of the pairs 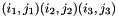 is different, and that difference is a transition.
is different, and that difference is a transition.
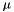 is a normalization factor.
is a normalization factor.
This model includes 2 parameters ( 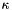 and
and 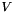 ). The codon frequencies
). The codon frequencies 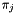 are either observed or infered.
are either observed or infered.
Reference:
- Goldman N. and Yang Z. (1994), Molecular Biology And Evolution 11(5) 725–736.
Constructor & Destructor Documentation
◆ GY94() [1/2]
| GY94::GY94 | ( | const GeneticCode * | gc, |
| FrequenciesSet * | codonFreqs | ||
| ) |
Definition at line 49 of file GY94.cpp.
References bpp::AbstractBiblioSubstitutionModel::lParPmodel_, bpp::AbstractBiblioSubstitutionModel::mapParNamesFromPmodel_, pmodel_, and bpp::AbstractBiblioSubstitutionModel::updateMatrices().
Referenced by clone().
◆ ~GY94()
◆ GY94() [2/2]
Member Function Documentation
◆ addRateParameter()
|
virtualinherited |
Implements bpp::SubstitutionModel.
Definition at line 84 of file AbstractBiblioSubstitutionModel.cpp.
References bpp::SubstitutionModel::addRateParameter(), bpp::AbstractBiblioSubstitutionModel::getModel(), bpp::AbstractBiblioSubstitutionModel::getRate(), bpp::AbstractBiblioSubstitutionModel::lParPmodel_, and bpp::AbstractBiblioSubstitutionModel::mapParNamesFromPmodel_.
◆ clone()
|
inlinevirtual |
Implements bpp::AbstractBiblioSubstitutionModel.
Definition at line 105 of file GY94.h.
References GY94().
◆ d2Pij_dt2()
|
inlinevirtualinherited |
- Returns
- The second order derivative of the probability of change from state i to state j with respect to time t, at time t.
- See also
- getd2Pij_dt2(), getStates()
Implements bpp::SubstitutionModel.
Definition at line 112 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::d2Pij_dt2(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ dPij_dt()
|
inlinevirtualinherited |
- Returns
- The first order derivative of the probability of change from state i to state j with respect to time t, at time t.
- See also
- getdPij_dt(), getStates()
Implements bpp::SubstitutionModel.
Definition at line 111 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::dPij_dt(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ enableEigenDecomposition() [1/2]
|
inlinevirtualinherited |
Set if eigenValues and Vectors must be computed.
Implements bpp::SubstitutionModel.
Definition at line 128 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::enableEigenDecomposition(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ enableEigenDecomposition() [2/2]
|
inlinevirtualinherited |
Tell if eigenValues and Vectors must be computed.
Implements bpp::SubstitutionModel.
Definition at line 130 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::enableEigenDecomposition(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ fireParameterChanged()
|
inlinevirtualinherited |
Tells the model that a parameter value has changed.
This updates the matrices consequently.
Definition at line 177 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::updateMatrices().
◆ freq()
|
inlinevirtualinherited |
- Returns
- Equilibrium frequency associated to character i.
- See also
- getFrequencies(), getStates()
Implements bpp::SubstitutionModel.
Definition at line 106 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::freq(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getAlphabet()
|
inlinevirtualinherited |
- Returns
- Get the alphabet associated to this model.
Implements bpp::SubstitutionModel.
Definition at line 153 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getAlphabet(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getAlphabetStateAsChar()
|
inlinevirtualinherited |
- Parameters
-
index The model state.
- Returns
- The corresponding alphabet state as character code.
Implements bpp::SubstitutionModel.
Definition at line 100 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getAlphabetStateAsChar(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getAlphabetStateAsInt()
|
inlinevirtualinherited |
- Parameters
-
index The model state.
- Returns
- The corresponding alphabet state as character code.
Implements bpp::SubstitutionModel.
Definition at line 98 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getAlphabetStateAsInt(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getAlphabetStates()
|
inlinevirtualinherited |
- Returns
- The alphabet states of each state of the model, as a vector of int codes.
- See also
- Alphabet
Implements bpp::SubstitutionModel.
Definition at line 94 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getAlphabetStates(), and bpp::AbstractBiblioSubstitutionModel::getModel().
Referenced by bpp::YNGKP_M1::YNGKP_M1(), bpp::YNGKP_M2::YNGKP_M2(), bpp::YNGKP_M3::YNGKP_M3(), bpp::YNGKP_M7::YNGKP_M7(), and bpp::YNGKP_M8::YNGKP_M8().
◆ getColumnRightEigenVectors()
|
inlinevirtualinherited |
- Returns
- A matrix with right eigen vectors. Each column in the matrix stands for an eigen vector.
Implements bpp::SubstitutionModel.
Definition at line 141 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getColumnRightEigenVectors(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getd2Pij_dt2()
|
inlinevirtualinherited |
- Returns
- All second order derivatives of the probability of change from state i to state j with respect to time t, at time t.
- See also
- d2Pij_dt2()
Implements bpp::SubstitutionModel.
Definition at line 126 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getd2Pij_dt2(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getdPij_dt()
|
inlinevirtualinherited |
- Returns
- Get all first order derivatives of the probability of change from state i to state j with respect to time t, at time t.
- See also
- dPij_dt()
Implements bpp::SubstitutionModel.
Definition at line 124 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getdPij_dt(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getEigenValues()
|
inlinevirtualinherited |
- Returns
- A vector with all real parts of the eigen values of the generator of this model;
Implements bpp::SubstitutionModel.
Definition at line 136 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getEigenValues(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getExchangeabilityMatrix()
|
inlinevirtualinherited |
- Returns
- The matrix of exchangeability terms. It is recommended that exchangeability matrix be normalized so that the normalized generator be obtained directly by the dot product
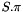 .
.
Implements bpp::SubstitutionModel.
Definition at line 118 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getExchangeabilityMatrix(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getFrequencies()
|
inlinevirtualinherited |
- Returns
- A vector of all equilibrium frequencies.
- See also
- freq()
Implements bpp::SubstitutionModel.
Definition at line 114 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getFrequencies(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getFrequenciesSet()
|
inlinevirtualinherited |
If the model owns a FrequenciesSet, returns a pointer to it, otherwise return 0.
Reimplemented from bpp::SubstitutionModel.
Reimplemented in bpp::YNGKP_M1, bpp::YNGKP_M8, bpp::YNGKP_M7, bpp::YNGKP_M3, and bpp::YNGKP_M2.
Definition at line 159 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getFrequenciesSet(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getGenerator()
|
inlinevirtualinherited |
- Returns
- The normalized Markov generator matrix, i.e. all normalized rates of changes from state i to state j. The generator is normalized so that (i)
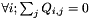 , meaning that $
, meaning that $ 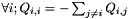 , and (ii)
, and (ii) 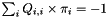 . This means that, under normalization, the mean rate of replacement at equilibrium is 1 and that time
. This means that, under normalization, the mean rate of replacement at equilibrium is 1 and that time 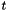 are measured in units of expected number of changes per site. Additionnaly, the rate_ attibute provides the possibility to increase or decrease this mean rate.
are measured in units of expected number of changes per site. Additionnaly, the rate_ attibute provides the possibility to increase or decrease this mean rate.
See Kosiol and Goldman (2005), Molecular Biology And Evolution 22(2) 193-9.
- See also
- Qij()
Implements bpp::SubstitutionModel.
Definition at line 116 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getGenerator(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getIEigenValues()
|
inlinevirtualinherited |
- Returns
- A vector with all imaginary parts of the eigen values of the generator of this model;
Implements bpp::SubstitutionModel.
Definition at line 138 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getIEigenValues(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getInitValue()
|
inlinevirtualinherited | ||||||||||||||||||||
This method is used to initialize likelihoods in reccursions. It typically sends 1 if i = state, 0 otherwise, where i is one of the possible states of the alphabet allowed in the model and state is the observed state in the considered sequence/site.
- Parameters
-
i the index of the state in the model. state An observed state in the sequence/site.
- Returns
- 1 or 0 depending if the two states are compatible.
- Exceptions
-
IndexOutOfBoundsException if array position is out of range. BadIntException if states are not allowed in the associated alphabet.
- See also
- getStates();
Implements bpp::SubstitutionModel.
Definition at line 157 of file AbstractBiblioSubstitutionModel.h.
References bpp::SubstitutionModel::getInitValue(), and bpp::AbstractBiblioSubstitutionModel::getModel().
◆ getModel() [1/2]
|
inlinevirtual |
Implements bpp::AbstractBiblioSubstitutionModel.
Definition at line 111 of file GY94.h.
References pmodel_.
◆ getModel() [2/2]
|
inlineprivatevirtual |
Implements bpp::AbstractBiblioSubstitutionModel.
Definition at line 114 of file GY94.h.
References pmodel_.
◆ getModelStates() [1/2]
|
inlinevirtualinherited |
Get the state in the model corresponding to a particular state in the alphabet.
- Parameters
-
code The alphabet state to check.
- Returns
- A vector of indices of model states.
Implements bpp::SubstitutionModel.
Definition at line 102 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::getModelStates().
◆ getModelStates() [2/2]
|
inlinevirtualinherited |
Get the state in the model corresponding to a particular state in the alphabet.
- Parameters
-
code The alphabet state to check.
- Returns
- A vector of indices of model states.
Implements bpp::SubstitutionModel.
Definition at line 104 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::getModelStates().
◆ getName()
|
inlinevirtual |
◆ getNumberOfStates()
|
inlinevirtualinherited |
Get the number of states.
For most models, this equals the size of the alphabet.
- See also
- getAlphabetChars for the list of supported states.
- Returns
- The number of different states in the model.
Implements bpp::SubstitutionModel.
Definition at line 155 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::getNumberOfStates().
◆ getPij_t()
|
inlinevirtualinherited |
- Returns
- All probabilities of change from state i to state j during time t.
- See also
- Pij_t()
Implements bpp::SubstitutionModel.
Definition at line 122 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::getPij_t().
◆ getRate()
|
inlinevirtualinherited |
Get the rate.
Implements bpp::SubstitutionModel.
Definition at line 143 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::getRate().
Referenced by bpp::AbstractBiblioSubstitutionModel::addRateParameter(), bpp::LG10_EX_EHO::LG10_EX_EHO(), bpp::LGL08_CAT::LGL08_CAT(), bpp::LLG08_EHO::LLG08_EHO(), bpp::LLG08_EX2::LLG08_EX2(), bpp::LLG08_EX3::LLG08_EX3(), bpp::LLG08_UL2::LLG08_UL2(), and bpp::LLG08_UL3::LLG08_UL3().
◆ getRowLeftEigenVectors()
|
inlinevirtualinherited |
- Returns
- A matrix with left eigen vectors. Each row in the matrix stands for an eigen vector.
Implements bpp::SubstitutionModel.
Definition at line 140 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::getRowLeftEigenVectors().
◆ getScale()
|
inlinevirtualinherited |
Get the scalar product of diagonal elements of the generator and the frequencies vector. If the generator is normalized, then scale=1. Otherwise each element must be multiplied by 1/scale.
- Returns
- Minus the scalar product of diagonal elements and the frequencies vector.
Implements bpp::SubstitutionModel.
Definition at line 190 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::getScale().
◆ getStateMap()
|
inlinevirtualinherited |
- Returns
- The mapping of model states with alphabet states.
Implements bpp::SubstitutionModel.
Definition at line 96 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::getStateMap().
◆ isDiagonalizable()
|
inlinevirtualinherited |
- Returns
- True if the model is diagonalizable in R.
Implements bpp::SubstitutionModel.
Definition at line 132 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::isDiagonalizable().
◆ isNonSingular()
|
inlinevirtualinherited |
- Returns
- True is the model is non-singular.
Implements bpp::SubstitutionModel.
Definition at line 134 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::isNonSingular().
◆ operator=()
Definition at line 78 of file GY94.cpp.
References bpp::AbstractBiblioSubstitutionModel::operator=(), and pmodel_.
◆ Pij_t()
|
inlinevirtualinherited |
- Returns
- The probability of change from state i to state j during time t.
- See also
- getPij_t(), getStates()
Implements bpp::SubstitutionModel.
Definition at line 110 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::Pij_t().
◆ Qij()
|
inlinevirtualinherited |
- Returns
- The rate in the generator of change from state i to state j.
- See also
- getStates();
Implements bpp::SubstitutionModel.
Definition at line 108 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::Qij().
◆ setFreq()
|
virtualinherited |
Set equilibrium frequencies.
- Parameters
-
frequencies The map of the frequencies to use.
Reimplemented from bpp::SubstitutionModel.
Definition at line 95 of file AbstractBiblioSubstitutionModel.cpp.
References bpp::AbstractBiblioSubstitutionModel::getModel(), bpp::AbstractBiblioSubstitutionModel::mapParNamesFromPmodel_, and bpp::SubstitutionModel::setFreq().
◆ setFreqFromData()
|
virtualinherited |
Set equilibrium frequencies equal to the frequencies estimated from the data.
- Parameters
-
data The sequences to use. pseudoCount A quantity 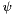 to add to adjust the observed values in order to prevent issues due to missing states on small data set. The corrected frequencies shall be computed as
to add to adjust the observed values in order to prevent issues due to missing states on small data set. The corrected frequencies shall be computed as ![\[ \pi_i = \frac{n_i+\psi}{\sum_j (f_j+\psi)} \]](form_280.png)
Implements bpp::SubstitutionModel.
Definition at line 110 of file AbstractBiblioSubstitutionModel.cpp.
References bpp::AbstractBiblioSubstitutionModel::getModel(), bpp::AbstractBiblioSubstitutionModel::mapParNamesFromPmodel_, and bpp::SubstitutionModel::setFreqFromData().
◆ setRate()
|
inlinevirtualinherited |
Set the rate of the model (must be positive).
- Parameters
-
rate must be positive.
Implements bpp::SubstitutionModel.
Definition at line 145 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::setRate().
◆ setScale()
|
inlinevirtualinherited |
Multiplies the current generator by the given scale.
- Parameters
-
scale the scale by which the generator is multiplied.
Implements bpp::SubstitutionModel.
Definition at line 192 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::setScale().
◆ Sij()
|
inlinevirtualinherited |
- Returns
- The exchangeability between state i and state j.
By definition Sij(i,j) = Sij(j,i).
Implements bpp::SubstitutionModel.
Definition at line 120 of file AbstractBiblioSubstitutionModel.h.
References bpp::AbstractBiblioSubstitutionModel::getModel(), and bpp::SubstitutionModel::Sij().
◆ updateMatrices()
|
protectedvirtualinherited |
Reimplemented in bpp::YNGKP_M1, bpp::YNGKP_M8, bpp::YNGKP_M7, bpp::YNGKP_M3, and bpp::YNGKP_M2.
Definition at line 70 of file AbstractBiblioSubstitutionModel.cpp.
References bpp::AbstractBiblioSubstitutionModel::getModel(), bpp::SubstitutionModel::getName(), bpp::AbstractBiblioSubstitutionModel::lParPmodel_, and bpp::AbstractBiblioSubstitutionModel::mapParNamesFromPmodel_.
Referenced by bpp::AbstractBiblioSubstitutionModel::fireParameterChanged(), GY94(), bpp::LG10_EX_EHO::LG10_EX_EHO(), bpp::LGL08_CAT::LGL08_CAT(), bpp::LLG08_EHO::LLG08_EHO(), bpp::LLG08_EX2::LLG08_EX2(), bpp::LLG08_EX3::LLG08_EX3(), bpp::LLG08_UL2::LLG08_UL2(), bpp::LLG08_UL3::LLG08_UL3(), bpp::MG94::MG94(), bpp::YNGKP_M2::updateMatrices(), bpp::YNGKP_M7::updateMatrices(), bpp::YNGKP_M8::updateMatrices(), bpp::YNGKP_M1::updateMatrices(), and bpp::YN98::YN98().
Member Data Documentation
◆ gacd_
◆ lParPmodel_
|
protectedinherited |
Definition at line 70 of file AbstractBiblioSubstitutionModel.h.
Referenced by bpp::AbstractBiblioSubstitutionModel::addRateParameter(), GY94(), bpp::LG10_EX_EHO::LG10_EX_EHO(), bpp::LGL08_CAT::LGL08_CAT(), bpp::LLG08_EHO::LLG08_EHO(), bpp::LLG08_EX2::LLG08_EX2(), bpp::LLG08_EX3::LLG08_EX3(), bpp::LLG08_UL2::LLG08_UL2(), bpp::LLG08_UL3::LLG08_UL3(), bpp::MG94::MG94(), bpp::AbstractBiblioSubstitutionModel::operator=(), bpp::YNGKP_M3::updateMatrices(), bpp::AbstractBiblioSubstitutionModel::updateMatrices(), bpp::YN98::YN98(), bpp::YNGKP_M1::YNGKP_M1(), bpp::YNGKP_M2::YNGKP_M2(), bpp::YNGKP_M3::YNGKP_M3(), bpp::YNGKP_M7::YNGKP_M7(), and bpp::YNGKP_M8::YNGKP_M8().
◆ mapParNamesFromPmodel_
|
protectedinherited |
Tools to make the link between the Parameters of the object and those of pmixmodel_.
Definition at line 68 of file AbstractBiblioSubstitutionModel.h.
Referenced by bpp::AbstractBiblioSubstitutionModel::addRateParameter(), GY94(), bpp::LG10_EX_EHO::LG10_EX_EHO(), bpp::LGL08_CAT::LGL08_CAT(), bpp::LLG08_EHO::LLG08_EHO(), bpp::LLG08_EX2::LLG08_EX2(), bpp::LLG08_EX3::LLG08_EX3(), bpp::LLG08_UL2::LLG08_UL2(), bpp::LLG08_UL3::LLG08_UL3(), bpp::MG94::MG94(), bpp::AbstractBiblioSubstitutionModel::operator=(), bpp::AbstractBiblioSubstitutionModel::setFreq(), bpp::AbstractBiblioSubstitutionModel::setFreqFromData(), bpp::YNGKP_M3::updateMatrices(), bpp::AbstractBiblioSubstitutionModel::updateMatrices(), bpp::YN98::YN98(), bpp::YNGKP_M1::YNGKP_M1(), bpp::YNGKP_M2::YNGKP_M2(), bpp::YNGKP_M3::YNGKP_M3(), bpp::YNGKP_M7::YNGKP_M7(), and bpp::YNGKP_M8::YNGKP_M8().
◆ pmodel_
|
private |
Definition at line 89 of file GY94.h.
Referenced by getModel(), GY94(), and operator=().
The documentation for this class was generated from the following files: