fix balance command
Syntax
fix ID group-ID balance Nfreq thresh style args keyword args ...
ID, group-ID are documented in fix command
balance = style name of this fix command
Nfreq = perform dynamic load balancing every this many steps
thresh = imbalance threshhold that must be exceeded to perform a re-balance
style = shift or rcb
shift args = dimstr Niter stopthresh dimstr = sequence of letters containing "x" or "y" or "z", each not more than once Niter = # of times to iterate within each dimension of dimstr sequence stopthresh = stop balancing when this imbalance threshhold is reached rcb args = none
zero or more keyword/arg pairs may be appended
keyword = weight or out
weight style args = use weighted particle counts for the balancing style = group or neigh or time or var or store group args = Ngroup group1 weight1 group2 weight2 ... Ngroup = number of groups with assigned weights group1, group2, ... = group IDs weight1, weight2, ... = corresponding weight factors neigh factor = compute weight based on number of neighbors factor = scaling factor (> 0) time factor = compute weight based on time spend computing factor = scaling factor (> 0) var name = take weight from atom-style variable name = name of the atom-style variable store name = store weight in custom atom property defined by fix property/atom command name = atom property name (without d_ prefix) out arg = filename filename = write each processor's sub-domain to a file, at each re-balancing
Examples
fix 2 all balance 1000 1.05 shift x 10 1.05
fix 2 all balance 100 0.9 shift xy 20 1.1 out tmp.balance
fix 2 all balance 100 0.9 shift xy 20 1.1 weight group 3 substrate 3.0 solvent 1.0 solute 0.8 out tmp.balance
fix 2 all balance 100 1.0 shift x 10 1.1 weight time 0.8
fix 2 all balance 100 1.0 shift xy 5 1.1 weight var myweight weight neigh 0.6 weight store allweight
fix 2 all balance 1000 1.1 rcb
Description
This command adjusts the size and shape of processor sub-domains within the simulation box, to attempt to balance the number of particles and thus the computational cost (load) evenly across processors. The load balancing is “dynamic” in the sense that rebalancing is performed periodically during the simulation. To perform “static” balancing, before or between runs, see the balance command.
Load-balancing is typically most useful if the particles in the simulation box have a spatially-varying density distribution or where the computational cost varies signficantly between different atoms. E.g. a model of a vapor/liquid interface, or a solid with an irregular-shaped geometry containing void regions, or hybrid pair style simulations which combine pair styles with different computational cost. In these cases, the LAMMPS default of dividing the simulation box volume into a regular-spaced grid of 3d bricks, with one equal-volume sub-domain per procesor, may assign numbers of particles per processor in a way that the computational effort varies significantly. This can lead to poor performance when the simulation is run in parallel.
The balancing can be performed with or without per-particle weighting. With no weighting, the balancing attempts to assign an equal number of particles to each processor. With weighting, the balancing attempts to assign an equal aggregate computational weight to each processor, which typically inducces a diffrent number of atoms assigned to each processor.
Note
The weighting options listed above are documented with the balance command in this section of the balance command doc page. That section describes the various weighting options and gives a few examples of how they can be used. The weighting options are the same for both the fix balance and balance commands.
Note that the processors command allows some control over how the box volume is split across processors. Specifically, for a Px by Py by Pz grid of processors, it allows choice of Px, Py, and Pz, subject to the constraint that Px * Py * Pz = P, the total number of processors. This is sufficient to achieve good load-balance for some problems on some processor counts. However, all the processor sub-domains will still have the same shape and same volume.
On a particular timestep, a load-balancing operation is only performed if the current “imbalance factor” in particles owned by each processor exceeds the specified thresh parameter. The imbalance factor is defined as the maximum number of particles (or weight) owned by any processor, divided by the average number of particles (or weight) per processor. Thus an imbalance factor of 1.0 is perfect balance.
As an example, for 10000 particles running on 10 processors, if the most heavily loaded processor has 1200 particles, then the factor is 1.2, meaning there is a 20% imbalance. Note that re-balances can be forced even if the current balance is perfect (1.0) be specifying a thresh < 1.0.
Note
This command attempts to minimize the imbalance factor, as defined above. But depending on the method a perfect balance (1.0) may not be achieved. For example, “grid” methods (defined below) that create a logical 3d grid cannot achieve perfect balance for many irregular distributions of particles. Likewise, if a portion of the system is a perfect lattice, e.g. the initial system is generated by the create_atoms command, then “grid” methods may be unable to achieve exact balance. This is because entire lattice planes will be owned or not owned by a single processor.
Note
The imbalance factor is also an estimate of the maximum speed-up you can hope to achieve by running a perfectly balanced simulation versus an imbalanced one. In the example above, the 10000 particle simulation could run up to 20% faster if it were perfectly balanced, versus when imbalanced. However, computational cost is not strictly proportional to particle count, and changing the relative size and shape of processor sub-domains may lead to additional computational and communication overheads, e.g. in the PPPM solver used via the kspace_style command. Thus you should benchmark the run times of a simulation before and after balancing.
The method used to perform a load balance is specified by one of the listed styles, which are described in detail below. There are 2 kinds of styles.
The shift style is a “grid” method which produces a logical 3d grid of processors. It operates by changing the cutting planes (or lines) between processors in 3d (or 2d), to adjust the volume (area in 2d) assigned to each processor, as in the following 2d diagram where processor sub-domains are shown and atoms are colored by the processor that owns them. The leftmost diagram is the default partitioning of the simulation box across processors (one sub-box for each of 16 processors); the middle diagram is after a “grid” method has been applied.

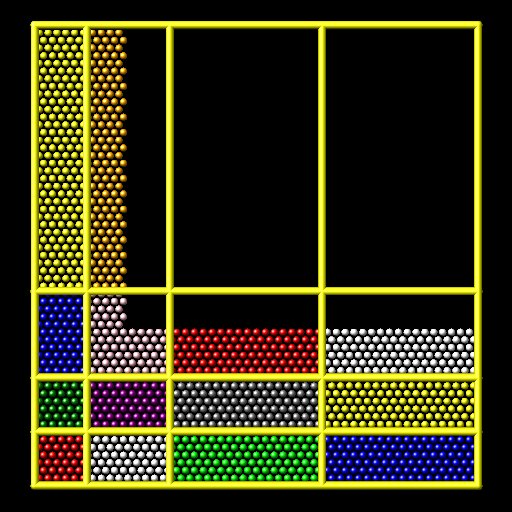

The rcb style is a “tiling” method which does not produce a logical 3d grid of processors. Rather it tiles the simulation domain with rectangular sub-boxes of varying size and shape in an irregular fashion so as to have equal numbers of particles (or weight) in each sub-box, as in the rightmost diagram above.
The “grid” methods can be used with either of the comm_style command options, brick or tiled. The “tiling” methods can only be used with comm_style tiled.
When a “grid” method is specified, the current domain partitioning can be either a logical 3d grid or a tiled partitioning. In the former case, the current logical 3d grid is used as a starting point and changes are made to improve the imbalance factor. In the latter case, the tiled partitioning is discarded and a logical 3d grid is created with uniform spacing in all dimensions. This is the starting point for the balancing operation.
When a “tiling” method is specified, the current domain partitioning (“grid” or “tiled”) is ignored, and a new partitioning is computed from scratch.
The group-ID is ignored. However the impact of balancing on different groups of atoms can be affected by using the group weight style as described below.
The Nfreq setting determines how often a rebalance is performed. If Nfreq > 0, then rebalancing will occur every Nfreq steps. Each time a rebalance occurs, a reneighboring is triggered, so Nfreq should not be too small. If Nfreq = 0, then rebalancing will be done every time reneighboring normally occurs, as determined by the the neighbor and neigh_modify command settings.
On rebalance steps, rebalancing will only be attempted if the current imbalance factor, as defined above, exceeds the thresh setting.
The shift style invokes a “grid” method for balancing, as described above. It changes the positions of cutting planes between processors in an iterative fashion, seeking to reduce the imbalance factor.
The dimstr argument is a string of characters, each of which must be an “x” or “y” or “z”. Eacn character can appear zero or one time, since there is no advantage to balancing on a dimension more than once. You should normally only list dimensions where you expect there to be a density variation in the particles.
Balancing proceeds by adjusting the cutting planes in each of the dimensions listed in dimstr, one dimension at a time. For a single dimension, the balancing operation (described below) is iterated on up to Niter times. After each dimension finishes, the imbalance factor is re-computed, and the balancing operation halts if the stopthresh criterion is met.
A rebalance operation in a single dimension is performed using a density-dependent recursive multisectioning algorithm, where the position of each cutting plane (line in 2d) in the dimension is adjusted independently. This is similar to a recursive bisectioning for a single value, except that the bounds used for each bisectioning take advantage of information from neighboring cuts if possible, as well as counts of particles at the bounds on either side of each cuts, which themselves were cuts in previous iterations. The latter is used to infer a density of pariticles near each of the current cuts. At each iteration, the count of particles on either side of each plane is tallied. If the counts do not match the target value for the plane, the position of the cut is adjusted based on the local density. The low and high bounds are adjusted on each iteration, using new count information, so that they become closer together over time. Thus as the recursion progresses, the count of particles on either side of the plane gets closer to the target value.
The density-dependent part of this algorithm is often an advantage when you rebalance a system that is already nearly balanced. It typically converges more quickly than the geometric bisectioning algorithm used by the balance command. However, if can be a disadvantage if you attempt to rebalance a system that is far from balanced, and converge more slowly. In this case you probably want to use the balance command before starting a run, so that you begin the run with a balanced system.
Once the rebalancing is complete and final processor sub-domains assigned, particles migrate to their new owning processor as part of the normal reneighboring procedure.
Note
At each rebalance operation, the bisectioning for each cutting plane (line in 2d) typcially starts with low and high bounds separated by the extent of a processor’s sub-domain in one dimension. The size of this bracketing region shrinks based on the local density, as described above, which should typically be 1/2 or more every iteration. Thus if Niter is specified as 10, the cutting plane will typically be positioned to better than 1 part in 1000 accuracy (relative to the perfect target position). For Niter = 20, it will be accurate to better than 1 part in a million. Thus there is no need to set Niter to a large value. This is especially true if you are rebalancing often enough that each time you expect only an incremental adjustement in the cutting planes is necessary. LAMMPS will check if the threshold accuracy is reached (in a dimension) is less iterations than Niter and exit early.
The rcb style invokes a “tiled” method for balancing, as described above. It performs a recursive coordinate bisectioning (RCB) of the simulation domain. The basic idea is as follows.
The simulation domain is cut into 2 boxes by an axis-aligned cut in the longest dimension, leaving one new box on either side of the cut. All the processors are also partitioned into 2 groups, half assigned to the box on the lower side of the cut, and half to the box on the upper side. (If the processor count is odd, one side gets an extra processor.) The cut is positioned so that the number of atoms in the lower box is exactly the number that the processors assigned to that box should own for load balance to be perfect. This also makes load balance for the upper box perfect. The positioning is done iteratively, by a bisectioning method. Note that counting atoms on either side of the cut requires communication between all processors at each iteration.
That is the procedure for the first cut. Subsequent cuts are made recursively, in exactly the same manner. The subset of processors assigned to each box make a new cut in the longest dimension of that box, splitting the box, the subset of processsors, and the atoms in the box in two. The recursion continues until every processor is assigned a sub-box of the entire simulation domain, and owns the atoms in that sub-box.
The out keyword writes text to the specified filename with the results of each rebalancing operation. The file contains the bounds of the sub-domain for each processor after the balancing operation completes. The format of the file is compatible with the Pizza.py mdump tool which has support for manipulating and visualizing mesh files. An example is shown here for a balancing by 4 processors for a 2d problem:
ITEM: TIMESTEP
0
ITEM: NUMBER OF NODES
16
ITEM: BOX BOUNDS
0 10
0 10
0 10
ITEM: NODES
1 1 0 0 0
2 1 5 0 0
3 1 5 5 0
4 1 0 5 0
5 1 5 0 0
6 1 10 0 0
7 1 10 5 0
8 1 5 5 0
9 1 0 5 0
10 1 5 5 0
11 1 5 10 0
12 1 10 5 0
13 1 5 5 0
14 1 10 5 0
15 1 10 10 0
16 1 5 10 0
ITEM: TIMESTEP
0
ITEM: NUMBER OF SQUARES
4
ITEM: SQUARES
1 1 1 2 3 4
2 1 5 6 7 8
3 1 9 10 11 12
4 1 13 14 15 16
The coordinates of all the vertices are listed in the NODES section, 5 per processor. Note that the 4 sub-domains share vertices, so there will be duplicate nodes in the list.
The “SQUARES” section lists the node IDs of the 4 vertices in a rectangle for each processor (1 to 4).
For a 3d problem, the syntax is similar with 8 vertices listed for each processor, instead of 4, and “SQUARES” replaced by “CUBES”.
Restart, fix_modify, output, run start/stop, minimize info:
No information about this fix is written to binary restart files. None of the fix_modify options are relevant to this fix.
This fix computes a global scalar which is the imbalance factor after the most recent rebalance and a global vector of length 3 with additional information about the most recent rebalancing. The 3 values in the vector are as follows:
- 1 = max # of particles per processor
- 2 = total # iterations performed in last rebalance
- 3 = imbalance factor right before the last rebalance was performed
As explained above, the imbalance factor is the ratio of the maximum number of particles (or total weight) on any processor to the average number of particles (or total weight) per processor.
These quantities can be accessed by various output commands. The scalar and vector values calculated by this fix are “intensive”.
No parameter of this fix can be used with the start/stop keywords of the run command. This fix is not invoked during energy minimization.
Restrictions
For 2d simulations, the z style cannot be used. Nor can a “z” appear in dimstr for the shift style.